Pain points in Writing Original Research Articles
October 11, 2022
Tips for Scholars to Improve Academic Writing Skills
November 21, 2022In-Brief
A wide range of scientific topics were covered, with bioinformatics receiving significant attention due to its rapid growth and growing necessity in biological data processing, particularly for huge omics datasets.
- Bioinformatics refers to using information technology and computers to huge molecular biology data sets.
- Bioinformatics is expected to be a cutting-edge part of the biotechnology sector that will aid medication development and individualized medical treatments.
- Artificial intelligence and computer science are strongly related to microbiology and genetics in this subject.
Introduction
Computer biology and bioinformatics are multidisciplinary fields that create and use computational methods to analyze massive quantities of biological data, such as genetic sequences, cell populations, or protein samples, to make predictions or find novel biology. Analytical approaches, mathematical modelling, and simulation are the computational methods employed.
In bioinformatics and computational biology, extracting intrinsic meaningful knowledge from huge omics data remains a formidable task. Deep learning, an emerging field of machine learning, has demonstrated exceptional performance in various academic and industrial applications. We emphasize the differences and similarities in frequently used deep learning models by addressing their basic architecture and examining different uses and drawbacks. We anticipate that the study will be useful in further developing its theory, method, and application in bioinformatics and computational biology.
Systematic review writing
Bioinformatics fosters opportunities for discoveries and offers avenues for biological investigations through data analysis. To provide exact results, Research writing a systematic review on this topic relies heavily on high-quality databases. However, most biological databases contain easily accessible errors, such as incorrectly categorized data or insufficient information. These errors might be serious. Recent data mining methods may use the filter data. However, these algorithms occasionally cannot cure these errors, resulting in serious analytical issues. A solution to this challenge is manual data curation and data extraction from books. A database of mutation effects on protein-ligand affinities, for example, was created after a thorough review of the literature. Nonetheless, the manual analysis may address the danger of bias.
Manual curation also allows for detecting errors and, where feasible, their correction. As a result, systematic literature reviews were employed to gather data for bioinformatics analysis. A systematic review is a strategy for discovering, estimating, and summarizing the state-of-the-art of a certain topic in the literature. Systematic review writing helps in the constrained gathering of literature material, allowing for a thorough methodological examination with less bias than typical reviews. A systematic review aims to develop a comprehensive picture of a specific subject and provide a good literature summary. Following a pre-established and well-defined process while conducting systematic reviews is critical.
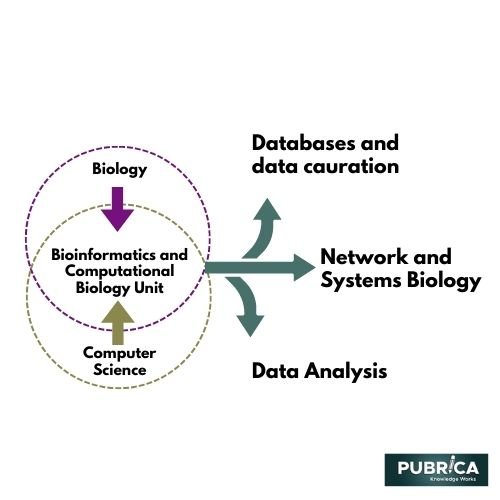
Computing in Bioinformatics and Computational Biology:
Bioinformatics and computational biology are emerging sciences that have recently made conducting a systematic review wave in technology sectors and the media. They are two of the few disciplines that need a broad range of expertise, from biology to computing. Wikipedia defines bioinformatics as “applying techniques from applied mathematics, informatics, statistics, computer science, chemistry, and biochemistry to address biological issues, generally at the molecular level.”
Numerous articles and essays on the subject have been produced, but locating excellent ones is like hunting for needles in a haystack. This issue is addressed in Parallel Computing for Bioinformatics and Computational Biology, a meticulous compilation of 29 articles and publications. The articles were divided into five categories: algorithms, sequence analysis, phylogenetics, protein folding, and technological platforms.
Structural bioinformatics: molecular folding, modelling, and design
One of the most common bioinformatics applications clinical trial systematic review services is the identification of three-dimensional structure, protein molecular modelling, and folding to forecast the possible function of proteins or model the behaviour of molecules, and other molecular structures, fold the molecule to its natural biologically functional three-dimensional structure and assist in the development of biomedical drugs for various complicated human diseases.
Biological networks and system biology
Bioinformatics is one of several scientific disciplines where the principles of network topology have found extensive application. As a result, large-scale biological networks, such as the biome, interactome, and microbiome, have been created.
Software, analysis tools, services, and workflow
The primary force behind the current and future advancement of bioinformatics tools and software is the advancement of genome decoding technologies, which is necessary for their analyses, the accumulation of large volumes of biological data, and the development of computer technologies, including networking, visualization, graphics, and molecular modelling.
Text mining
The increasing volume of biomedical literature is being gathered, constructed, and organized using computer algorithms and bioinformatics tools. This encourages scientists to query, mine, examine, and synthesize the specific literature and published articles of their research interest.
Conclusion
Systematic literature reviews were done to gather information for bioinformatics analysis. It is a method for determining, estimating, and summarizing the current status of a certain literary topic. In contrast to traditional reviews, systematic literature reviews allow for the selective collection of literature material, enabling a rigorous methodological analysis with less prejudice. The objective is to understand a particular issue and provide a fair assessment of the literature. As a result, adhering to a pre-established and well-stated process while conducting systematic reviews is crucial.
About Pubrica
Scientific and medical research papers are produced by the team of researchers and writers at Pubrica, and they may be invaluable sources for authors and practitioners. Pubrica medical writers help you create and modify the introduction by using the reader to alert them to the gaps in the selected study subject. Our experts know the sequence in which the topic where the hypothesis is given is followed by the broad subject, the issue, and the backdrop.
References
- Fu, Yuanyuan, et al. “Current trend and development in bioinformatics research.” BMC Bioinformatics 21.9 (2020): 1-3.
- Zomaya, A. Y. (2005). Parallel computing for bioinformatics and computational biology. Wiley.
- Vega-Rodríguez MA, Rubio-Largo Á. Parallelism in computational biology: A view from diverse high-performance computing applications. The International Journal of High Performance Computing Applications. 2018;32(3):317-320. doi:10.1177/1094342016677599