The Importance of Case Report Writing in Evidence-based Practice
February 24, 2021
Effective Strategies to Monitor Clinical Risks using Biostatistics
March 9, 2021Current Developments in Bioinformatics and Computational Biology
In-Brief
- Certain types of gene therapy help in improving the body’s ability to recognize and destroy cancer cells. Certain immune system cells are responsible for recognizing and killing these cells
- Certain types of gene therapy help in improving the body’s ability to recognize and destroy cancer cells. Certain immune system cells are responsible for recognizing and killing these cells
Introduction
Gene therapy is the process of altering the genes in your body’s cells to cure or prevent disease. Your DNA — the code that governs all of your body’s structure and function, from making you taller to controlling your body systems — is stored in your genes. The disease caused by genes that aren’t working properly.
Gene therapy is a treatment that comprises altering the genes in your body’s cells to treat or prevent disease. Your genes store your DNA, which is the code that controls all of the structure and function of your body, from making you taller to regulating your body systems. Genes that aren’t functioning properly can cause disease.
Why it’s done
Your genes store your DNA, which is the code that controls all of the structure and function of your body, from making you taller to regulating your body systems. Genes that aren’t functioning properly can cause disease research paper writing help in understanding the concept of gene therapy in detail.
- Replacing mutated genes. Since some genes function incorrectly or no longer function at all, certain cells become diseased. Replacement of defective genes can aid in the treatment of certain diseases. A gene called p53, for example, usually prevents tumour development. Problems with the p53 gene linked to a variety of cancers. If doctors could substitute the cancer cells’ faulty p53 gene, the cancer cells would die.
- Fixing mutated genes. Mutated genes that cause disease switched off to stop promoting disease, or stable genes that help avoid disease c turned on to stop the disease from spreading.
- Constructing diseased cells more evident to the immune system. Since your immune system doesn’t identify diseased cells as intruders in some cases, it doesn’t attack them. Specialists were able to use gene therapy to teach the immune system to recognize dangerous cells.
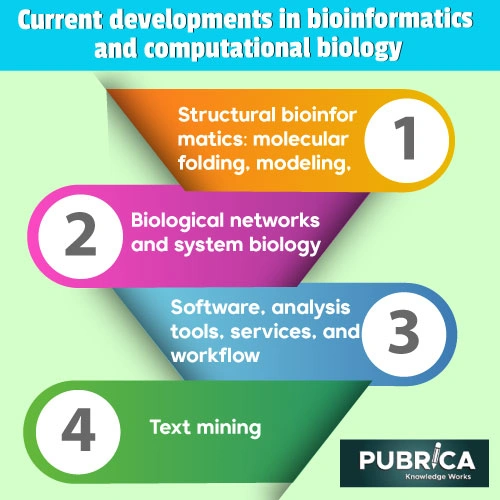
Biological networks and system biology
The properties of network topology are widely applied in many scientific fields, including bioinformatics, leading to the construction of large-scale biological networks represented as “omes” like biome, interactome, microbiome.
The above emphasized molecular sequence analysis, annotation, prediction, and molecular modelling-associated to bioinformatics methods are also the principal for organizing, building, and genetic and biochemical pathways of complex cellular processes. These signal transduction comprise reception, and gene co-expression and gene regulation. Such molecular networks incorporate various data types, including DNA sequences, regulatory RNA, proteins, gene expression data, secondary metabolites, and other small molecules, which may be connected physically and functionally. The structuring and organization of such physically and functionally connected molecular networks of cellular processes can be achieved only by applying the combination of simulative, iterative, and model-oriented bioinformatics approaches.
Software, analysis tools, services, and workflow
The key driving force for the present and future development of bioinformatics software and tools, which help in the progress of genome decoding technologies, consequent required for their analyses, accretion of large volume biological data, as well as the development of computer technologies, visualization, graphics, and networking techniques and molecular modelling.
Moreover, the availability of numerous shared object models, open-source codes, and community-maintained plug-ins helps collect new ideas from the community and perform innovative in silico experiments on current “Big Data.”
These all make way for research groups and bioinformatics corporations to experiment, work, and create a more innovative generation of bioinformatics tools and software that are user-friendly and must perform integrated and extended analysis with better visualization and graphical outputs.
Some of the open-source software packages include UGENE, EMBOSS, GenGIS, GENtle, BioPerl, PathVisio, GenoCAD, GenomeSpace, Biopython, Bioclipse, GeWorkbench, .NET Bio, Apache Taverna, BioJS, Bioconductor, BioJava, and BioRuby.
Text Mining
Bioinformatics research and application aim to use computational algorithms and bioinformatics tools to gather, construct, and structure the increasing body of biomedical literature permutes scientists to question, mine, study, and synthesize the particular literature and published articles of their research interest.
Therefore, text mining and biomedical literature play a significant role in the scientific enhancement, inventions, and application and integration of discoveries to society through extracting information (EI) and assessing the relationships of publications.
Biomedical literature text mining uses various “text mining & data mining” tools, visualization and navigation, information retrieval, applying techniques such as data clustering and extraction, and text categorization and summarization.
Conclusion
Systematic literature reviews used to collect data for bioinformatics analysis. It is a technique used to discover, estimate and summarize the state-of-the-art of a precise theme in the literature. Systematic Literature Review permits gathering literature information restrictively that allows a rigorous methodological analysis with lower bias than the traditional reviews. The objective is to build a general vision of a specific question and give it a reasonable summary of the literature. Hence, to perform systematic reviews is essential to follow a pre-established and well-defined protocol.
References
- Zomaya, A. Y. (2005). Parallel computing for bioinformatics and computational biology. Wiley.
- Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., … & Zhang, J. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biology, 5(10), 1-16.